Transparent and Reproducible Research with R (Part 1)
And a bit of git/GitHub
Daniel Anderson
Joshua M Rosenberg
April 7, 2019
Agenda
First two hours
- Introduction (8:00 - 8:45)
- Who we are, who participants are and why they're here (15 min.)
- Reproducible research and literate programming (20 min.)
- Conducting science in the public and in an open way & OSF (10 min.)
Agenda
First two hours
- Introduction (8:00 - 8:45)
- Who we are, who participants are and why they're here (15 min.)
- Reproducible research and literate programming (20 min.)
- Conducting science in the public and in an open way & OSF (10 min.)
- R Markdown (8:45 - 10:00)
- Delineating code chunks from plain text (15 min.)
- Creating headers (5 min.)
- Creating lists and using other features of markdown (10 min.)
- Whole-document and code chunk-specific options (15 min.)
- Rendering and sharing documents in different formats (15 min.)
- Lab (practice) (15 min.)
Break (10:00 - 10:15)
Last two hours
Advanced R Markdown functionality (10:15 - 11:00 minutes)
- Formatting tables (20 min.)
- Creating manuscripts to submit for publication (via {papaja}; 25 min.)
Use of git/GitHub for version control and collaboration (11:00 - 11:45)
- Introduction to GitHub, RStudio interface, and GitKraken GUI (20 min.)
- Making changes, committing them, and pushing them to the repository (15 min.)
- Use of GitHub (and ignoring specific files via
.gitignore; 10 min.)
Wrap-up/ideas for next steps/staying in touch (11:45 - 12:00)
#whoami
- Research Assistant Professor: Behavioral Research and Teaching, University of Oregon (#goducks)
- Dad (two daughters: 6 (almost 7) and 4)
- Primary areas of interest
- 💗💗R💗💗 and computational research
- Open data, open science, and reproducible workflows
- Growth modeling, achievement gaps, and variance between educational institutions (particularly spatially)

#whoami 2
- Assistant Professor: STEM Education, University of Tennessee, Knoxville
- Also a Dad (one-year-old toddler!)
- Primary areas of interest
- Data science in education (network analytic methods, experience sampling method, computational grounded theory)
- Data science education (integrating data science and science education)

#whoyouis
Introduce yourself What's your prior experience with R/R Markdown and git/GitHub? Why are you here?
This slide means we're
transitioning to Daniel
This slide means we're
transitioning to Josh
Before we really get started
Realize this is all going to be an intro
Covering a lot of content - that's purposeful
Idea is to give you exposure and some basic familiarity
Before we really get started
Realize this is all going to be an intro
Covering a lot of content - that's purposeful
Idea is to give you exposure and some basic familiarity
Resources to learn more
- Daniel's class(es)
- Intro course
- Data viz course
(plenty of others out there too)
Reproducible Research and
Literate Programming
8:15 - 8:30am
A couple caveats
- Much of what we're going to be discussing represents an ideal that we have only recently begun working towards.
A couple caveats
Much of what we're going to be discussing represents an ideal that we have only recently begun working towards.
None of what we will talk about should be taken as a referendum on you or your current practices. However, we hope to help to convince you that you should be working toward the reproducible research ideal, and that, as a field, we should be moving toward reproducible research being the minimal standard.
A couple caveats
Much of what we're going to be discussing represents an ideal that we have only recently begun working towards.
None of what we will talk about should be taken as a referendum on you or your current practices. However, we hope to help to convince you that you should be working toward the reproducible research ideal, and that, as a field, we should be moving toward reproducible research being the minimal standard.
We will be focusing on reproducible research with R (obviously). Other options are available but, in our view, none are as clear, comprehensive, and easy to implement as the tools at your disposal through R.
What is reproducible research?
- Replicability is the gold standard for research. Ideally, most research would be verified through replication.
What is reproducible research?
Replicability is the gold standard for research. Ideally, most research would be verified through replication.
Reproducibility represents a minimal standard, which itself can aid replication (tremendously), by conducting and documenting the research sufficiently that an independent researcher could reproduce all the results from a study, provided the data were available
What is reproducible research?
Replicability is the gold standard for research. Ideally, most research would be verified through replication.
Reproducibility represents a minimal standard, which itself can aid replication (tremendously), by conducting and documenting the research sufficiently that an independent researcher could reproduce all the results from a study, provided the data were available
Turns out this is a more difficult standard than we would generally like to admit.
Why should we care?
Reproducibility as an ethical standard
- More transparency
- More potential for results to be verified (and errors found/corrected)
Why should we care?
Reproducibility as an ethical standard
- More transparency
- More potential for results to be verified (and errors found/corrected)
If your work is not reproducible, it is often not truly replicable.
Why should we care?
Reproducibility as an ethical standard
- More transparency
- More potential for results to be verified (and errors found/corrected)
If your work is not reproducible, it is often not truly replicable.
If your work is reproducible, then others have a "recipe" for replication.
Are journal articles research?
- Initially, we may think of journal articles as research, but really the research is everything that went into the article, not the article itself.
Are journal articles research?
Initially, we may think of journal articles as research, but really the research is everything that went into the article, not the article itself.
Some (Buckheit & Donoho, 2015) conceive of the article as the "advertisement".
Are journal articles research?
Initially, we may think of journal articles as research, but really the research is everything that went into the article, not the article itself.
Some (Buckheit & Donoho, 2015) conceive of the article as the "advertisement".
- If all we have is the advertisement, can we really fully understand the
steps and decisions made during the research?
- In large-scale data analysis, the answer is generally "no".
Tangential benefits
Striving toward reproducible research will:
Make your own code more efficient/easily interpretable
- Can help with collaboration on a project
Reduce errors
Increase efficiency by not having to redo tables and figures with each tweak to a model.
What does literate programming look like?
What this workshop is about!
What does literate programming look like?
What this workshop is about!
- Start with a basic text document (not Word, text)
What does literate programming look like?
What this workshop is about!
Start with a basic text document (not Word, text)
Use the text document to write your article
What does literate programming look like?
What this workshop is about!
Start with a basic text document (not Word, text)
Use the text document to write your article
Embed code within the text document that corresponds to your analysis. Note this is not just copying the code in. The code should be live and what you're working with while conducting your research.
What does literate programming look like?
What this workshop is about!
Start with a basic text document (not Word, text)
Use the text document to write your article
Embed code within the text document that corresponds to your analysis. Note this is not just copying the code in. The code should be live and what you're working with while conducting your research.
Render the document into a different format (pdf, html, etc.).
- Select which code (if any) will be displayed
- Build tables of results and plots to be produced
End result
Readers can then read the "advertisement", but if they are interested in reproducing your results they can access the text file that contains the analysis code.
Single product that has the advertisement and the research process embedded.
Other reasons dynamic documents are useful
Outside of reproducibility, you may want to use R Markdown to:
Produce slides
Keep track of your analysis (notes, essentially), even if you end up using something like Word
Share code with others
Quickly share results with others
Produce professional looking data products
Challenges
- Word is the industry standard (frustratingly so, to us)
- Word output is less than ideal
- Can be difficult when collaborating with others
- Some journal articles require papers submitted in Word
- Potentially get a pdf to word converter, but still less than ideal
- Advanced features have a relatively steep learning curve
Open science and R Markdown basics
8:35 - 8:45 am
Open science and public work
Benefits
- Working in the public has potential benefits:
- People know what your expertise is
- Potential colleagues can reach out to you for collaborations
- Allow others to build upon your work
- Build a network
Dilemmas
- Working in the public has some potential dilemmas/drawbacks to manage:
- Sometimes difficult to share pre-prints (due to copyright issues)
- Getting 'scooped'
- Often difficult to share data
Open Science Framework
Offers a platform in which you can store private data
Has an Application Programming Interface (API) to connect with R
Example: https://osf.io/9ex7k/
R Markdown Basics
8:45 - 9:00
R Markdown
From within your R Studio Project:
First thing: Render!
Create new a R Markdown doc
### Try it out!
YAML Front Matter
---title: Example Markdown documentauthor: - Daniel Anderson - : "2019-05-07"---
- Three dashes before and after the YAML fields
- Case sensitive
- Many other fields are possible.
- For example, you may want to include an
output:argument (pdf_document,html_document,word_document). Must be specified as it is rendered, if not supplied.
- For example, you may want to include an
Example: Change syntax highlighting
The YAML will control a lot of how a document looks. For example, if you wanted to render with a different syntax highlighter:
Standard Rmd
---title: "Doc Title"output: pdf_document---kate
---title: "Doc Title"output: pdf_document: highlight: kate---Code chunks versus text
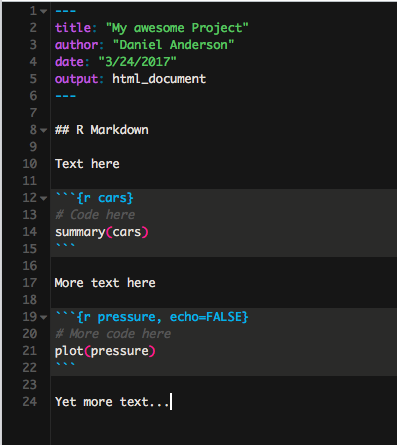
Code chunks
Start a code chunk with ```{r}, then produce some r code, then close the
chunk with three additional back ticks ``` .

Code chunks
Start a code chunk with ```{r}, then produce some r code, then close the
chunk with three additional back ticks ``` .

a <- 3b <- 5a + b * (exp(a)/b)## [1] 23.08554R Markdown basics
Headings, lists, chunk options, and inline code
9:00 - 9:30am
Headings and Lists
Not R-lists
# Level 1## Level 2 ### Level 3 (etc.) * Unordered list - inset + inset more - etc.1. Ordered list a. blah blah2. More stuff
echo and eval
You can show code without evaluating it, using eval = FALSE.

a + b * (exp(a)/b)Alternatively, you can evaluate the code without displaying it, using echo =
FALSE.

warning
Warning = FALSE
ggplot(msleep, aes(sleep_rem, sleep_total)) + geom_point() Warning is printed to the console when rendering.
Warning is printed to the console when rendering.
Warning = TRUE
ggplot(msleep, aes(sleep_rem, sleep_total)) + geom_point()## Warning: Removed 22 rows containing missing values (geom_point).
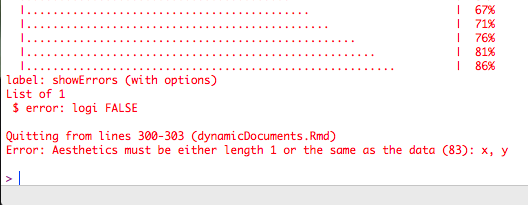
Show errors
error = TRUE
ggplot(msleep, aes(sleep, sleep_total)) + geom_point()## Error: Aesthetics must be either length 1 or the same as the data (83): xShow errors
error = TRUE
ggplot(msleep, aes(sleep, sleep_total)) + geom_point()## Error: Aesthetics must be either length 1 or the same as the data (83): xIf error = FALSE, the document won't render if it encounters an error.
Message
Some functions will return messages. You may want to suppress these.
message = FALSE
ggplot(msleep, aes(sleep_total)) + geom_histogram()
Message
message = TRUE
ggplot(msleep, aes(sleep_total)) + geom_histogram()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
include
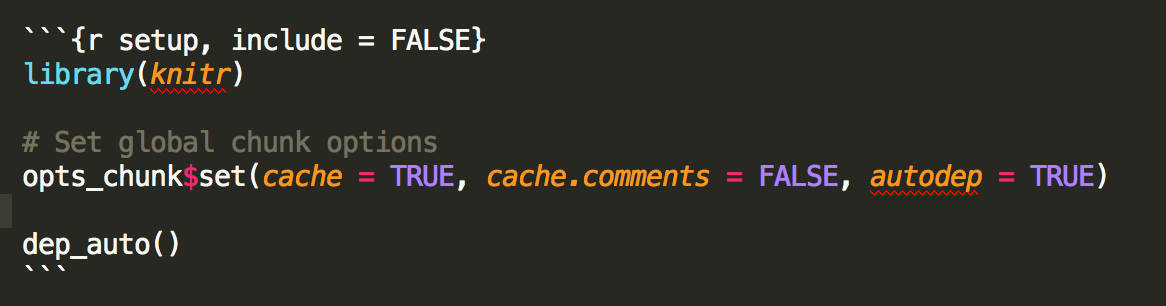
The include argument is used to evaluate code that is not included in the
document at all. For example, when setting up your global options.
Setting global options
Change the default behavior
opts_chunk$set(...) # insert options hereFor example, you can set echo = FALSE and fig.width = 6.5 and fig.height = 8 with the following code.
opts_chunk$set(echo = FALSE, fig.width = 6.5, fig.height = 8)This is most useful when producing a report for somebody who doesn't use R and has no use or knowledge of the code.
You can always override the global options within a particular chunk, e.g.
```{r, chunkName, echo = TRUE}
```
Other things to consider setting globally:
warnings = FALSEmessage = FALSEerrors = TRUEecho = FALSE- Caching options (next slides)
More complete chunk options
| Options | Arguments | Default | Result |
|---|---|---|---|
| eval | logical | TRUE | Evaluate the code? |
| echo | logical | TRUE | Show the code? |
| results | markup, asis, hold, hide | markup | Render the results |
| warning | logical | TRUE | Print warnings? |
| error | logical | TRUE | Preserve errors? (if FALSE, quit) |
| message | logical | TRUE | Print any messages? |
| include | logical | TRUE | Include any of the code or output or code? |
| tidy | logical | FALSE | Tidy code? (see formatR package) |
(and a few more)
| Options | Arguments | Default | Result | |
|---|---|---|---|---|
| 9 | cache | logical, 0:3 | FALSE | Cache code chunks? |
| 10 | cache.comments | logical | NULL | Cache invalidated by comment changes? |
| 11 | dependson | char, num | NULL | Current chunk depend on prior cached chunks? |
| 12 | autodep | logical | FALSE | Depends determined automatically? |
| 13 | fig.height/fig.width | numeric | 7, 7 | Height and width of figure |
| 14 | fig.show | asis, hold, animate, hide | asis | How the figure should be displayed |
| 15 | interval | numeric | 1 | Animate speed |
For complete documentation, see http://yihui.name/knitr/options/
Inline code
A single back tick followed by r produces inline code to be evaluated.

This is an example of inline code, where I want to refer to the sum of a and
b, which is 8.
This is extremely useful in writing reports. Never have to update any numbers in text, regardless of changes to your models or data (if you are careful about it).
Real example
Rendering and documents
9:30 - 10:00am-
Rendering R Markdown documents
Modify the YAML
Get the same document to render to different formats by modifying the YAML to output HTML, PDF, and .docx
From
---title: "My Document"author: "Stephanie Lawson"output: html_document---To
---title: "My Document"author: "Stephanie Lawson"output: pdf_document---Rendering to HTML
---title: "My Document"author: "Stephanie Lawson"output: html_document: toc: true toc_depth: 2 toc_float: true number_sections: true higlight: kate---Re# Rendering to a PDF
- Need a tex (pronounced tek) distribution
- Our recommendation for this workshop with probably everything you'll ever need: {tinytex}
Re# Rendering to a PDF
- Need a tex (pronounced tek) distribution
- Our recommendation for this workshop with probably everything you'll ever need: {tinytex}
install.packages("tinytex")tinytex::install_tinytex()This is another amazing package by Yihui Xie. See more about it here
ndering to PDF
---title: "My Document"author: "Stephanie Lawson"output: pdf_document: toc: true number_sections: true higlight: kate---Rendering to .docx
A key feature is the ability to use a reference document; see here
---title: "My Document"author: "Stephanie Lawson"output: word_document---